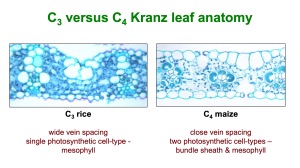
What is Kranz Anatomy?
In the majority of plants, including rice, CO2 is first fixed into a compound with three carbons (C3) by the photosynthetic enzyme ribulose bisphosphate carboxylase oxygenase (Rubisco) – this is known as C3 photosynthesis. Rubisco is inherently inefficient because it can also catalyze a reaction with oxygen, giving a wasteful process known as photorespiration (rather than photosynthesis). To overcome this inefficiency, the C4 pathway initially fixes atmospheric CO2 into C4 acids using the enzyme phosphoenolpyruvate carboxylase that is insensitive to O2. CO2 is then released from the C4 acids for re-fixation by Rubisco. In most C4 plants these two stages of the C4 pathway are spatially separated in morphologically distinct photosynthetic cell-types, allowing a high concentration of CO2 to accumulate in the vicinity of Rubisco, and greater photosynthetic efficiency.
In C4 grasses such as maize and some C4 dicots, enlarged bundle sheath (BS) cells surround the veins (V) and the BS cells are then surrounded by mesophyll (M) cells. Each pair of veins is thus separated by two BS and two M cells in a V-BS-M-M-BS-V pattern referred to as Kranz anatomy. The anatomy of C3 plants is typically a V-BS-M-M-M-M-M-M-M-M-BS-V pattern, and the BS cells are smaller than in C4 plants. A fully functional C4 pathway therefore requires a coordinated change in tissue structure and metabolic biochemistry. During evolution, these changes have occurred more than 50 times in a wide range of flowering plants, indicating that, despite being complex, it is a relatively easy pathway to evolve.
Towards an understanding of Kranz development
The development of Kranz anatomy can be considered in three general stages: 1) initiation of procambium; 2) BS and M cell-specification; 3) chloroplast development and the integration of the C4 cycle. To identify candidate regulators of procambium initiation and cell-type specification, we have compared genome-wide analyses of expression profiles in leaves of maize that either do (foliar) or do not (husk) develop Kranz anatomy. Our work has led to a model of Kranz regulation that is currently being tested. Ultimately this research will link to our earlier work on the regulation of chloroplast development in C4 plants (see below).
The role of GOLDEN2-like genes in chloroplast development
In C4 plants morphologically and biochemically distinct chloroplasts develop in BS and M cell-types. Over recent years, a great deal has been learnt about the photosynthetic apparatus, chloroplast protein import and many other aspects of chloroplast activity in the two cell-types. However, little is known of how the transition from a non-photosynthetic proplastid to a functional chloroplast is regulated. Our group defined a small family of genes encoding nuclear transcription factors that fits this role.
The GOLDEN2 gene was initially discovered in maize following analysis of the golden2 (g2) mutant (Hall et al., 1998; Langdale & Kidner, 1994). A GOLDEN2-LIKE (GLK) gene (ZmGLK1) was then identified in maize on the basis of homology to G2 (Rossini et al., 2001). Similar pairs of GLK genes were then identified in rice (Rossini et al., 2001) and Arabidopsis (Fitter et al., 2002).
The presence of duplicate genes in both C4 maize and C3 rice led us to propose that the two GLK genes act redundantly to promote the development of a single chloroplast type in C3 plants whereas each gene promotes development of one of the two specialized chloroplast types in C4 plants. We provided compelling support for this hypothesis by showing that GLK genes are also expressed cell-specifically in the C4 species Sorghum bicolor (Wang et al., 2013), and that single glk mutants in the C3 plants Arabidopsis and rice were phenotypically normal whereas double mutants exhibited impaired chloroplast development. In Arabidopsis, chloroplasts in the double mutant are small and possess a substantially reduced thylakoid network (below).
Further support for a specialized role for GLK genes in C4 plants came from a comprehensive phylogenetic analysis using whole genome data. This analysis demonstrated that GLK gene duplicates in C3 lineages can be traced back to whole genome duplications whereas those in C4 lineages are most likely to have resulted from gene-specific duplications (Wang et al., 2013). This suggests that gene duplications in C4 lineages may have been amongst some of the proposed pre-conditioning events for the evolution of C4.
We also showed that a pair of GLK genes are required for chloroplast development in the moss Physcomitrella patens, a representative of the most primitive extant land plants (Yasumura et al., 2005). Given that GLK genes are not found in any algal or non-plant genomes, these findings suggested that GLK transcription factors evolved to regulate chloroplast development as plants moved on to land.
To discover how GLK transcription factors regulate chloroplast development, we identified direct targets of GLK action. We showed that GLK genes interact with the promoters of a suite of genes encoding chlorophyll biosynthesis enzymes, light harvesting and electron transport components (Waters et al., 2009). These discoveries led us to propose that GLK proteins act to integrate environmental and developmental cues to maximize photosynthetic output, a scenario consistent with the suggestion that GLK function facilitated the adaptation of plants to land. This suggestion has now been supported by transcriptome and functional analyses in Arabidopsis thaliana and other flowering plants. These studies have shown that GLK proteins act at key points in brassinosteroid, circadian clock, fruit ripening, nitrogen-use and disease response pathways, all of which modify, or are modified by, the functional state of the chloroplast.
The C4 rice project
World rice production has been increasing during the last few decades. However, we will still need 25% more rice in 2025 to meet the growing needs of the human population. To further increase crop productivity, one of the options is to explore the possibility of transferring C4 traits into C3 plants.
In 2006, the International Rice Research Institute (IRRI) brought together a group of scientists from around the world to investigate the feasibility of developing C4 rice plants. The C4 consortium has been working together since 2008, and the C4 Rice project has just entered its third funding phase. We are applying our understanding of leaf anatomy and chloroplast development to the goal of generating C4 rice.